Cellartis iPSC rCas9 Electroporation and Single-Cell Cloning System User Manual
(030619)
takarabio.com
Takara Bio USA, Inc.
Page 10 of 24
3.
Purify your sgRNA after digestion with Recombinant DNase I (RNase-Free) using the Guide-it
IVT RNA Clean-Up Kit; measure its concentration using a NanoDrop 2000 spectrophotometer or
equivalent.
4.
If you wish to determine cleavage efficiency before proceeding with editing (Figure 4, gray
boxes), we recommend using the Guide-it sgRNA Screening Kit (sold separately).
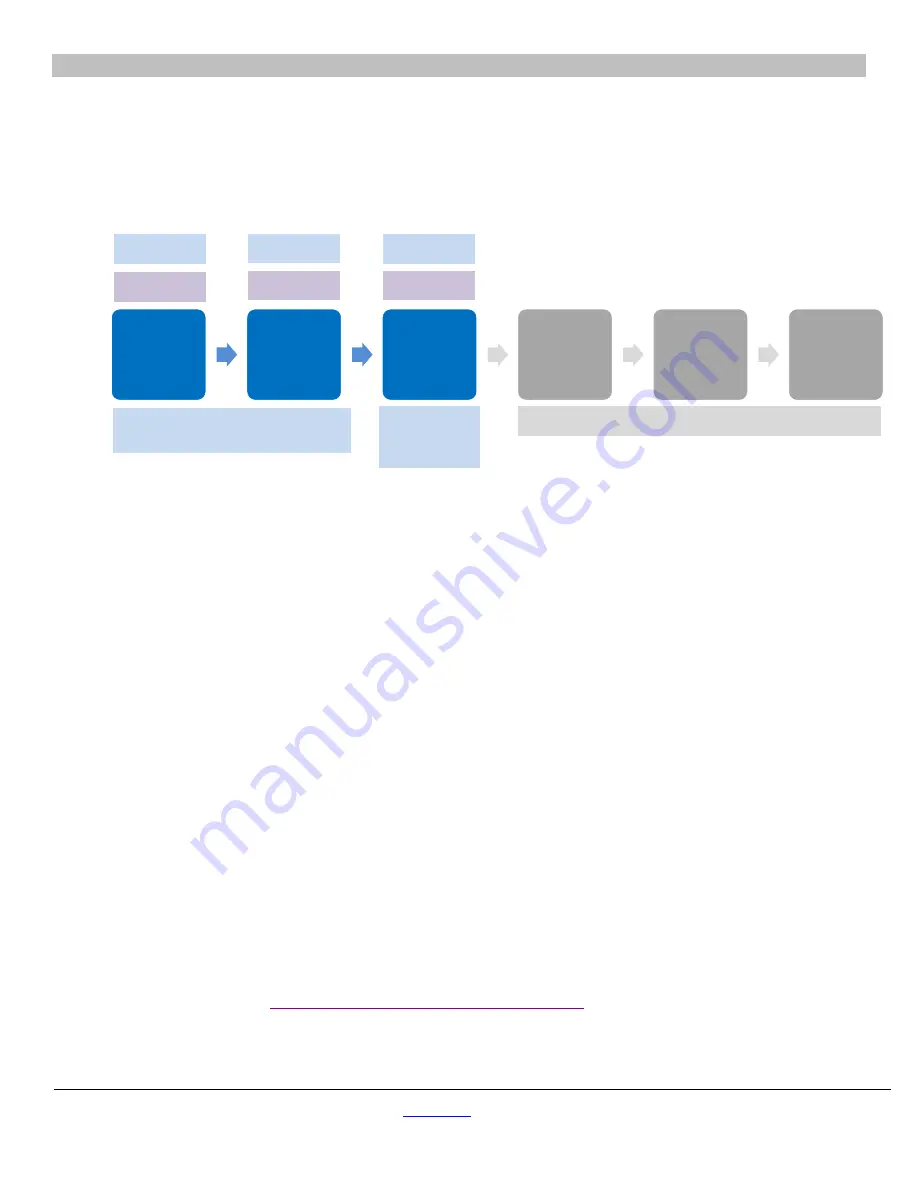
Figure 4. Workflow for generating sgRNA using components of the Cellartis iPSC rCas9 Electroporation and Single-Cell Cloning
System.
Steps 1–3 describe the workflow for using Guide-it sgRNA In Vitro Transcription Components v2 and the Guide-it IVT RNA
Clean-Up Kit to synthesize and purify sgRNAs. The purple boxes indicate the sections containing the relevant protocols. The gray boxes
indicate optional steps of
in vitro
sgRNA screening.
B.
Generating the DNA Template
Guidelines for Designing PCR Primers
Use the following guidelines to design a forward primer to be used in a PCR reaction with the included
Guide-it Scaffold Template to create a DNA template for
in vitro
transcription of your sgRNA. This
primer should contain the T7 promoter sequence, followed by your sgRNA target sequence, and the
Guide-it Scaffold Template-specific sequence (Figure 5).
Choosing the Correct DNA Target Sequence
Choose the DNA target sequence that will correspond to your actual sgRNA target sequence as
shown in Figure 5, Panel A, according to the following guidelines:
a.
The DNA target sequence you choose must end with the proto-spacer adjacent motif
(PAM) sequence, NGG, on its 3’ end. Only DNA sequences that are 20 nucleotides
upstream of a PAM sequence can be used for CRISPR/Cas9.
b.
Any target sequence can be used as long as the sequence is followed by the PAM
sequence, NGG. However, to minimize off-target cleavage events, the entire target
sequence (including the PAM) should have at least three base mismatches with any other
non-targeted genomic sequence. Off-target events should be especially low if the
mismatches are in, or adjacent to, the PAM. Most online tools for sgRNA design will
predict off-target sequences for a given sgRNA target sequence. To learn more, visit
http://www.takarabio.com/sgRNA-design-tools
.
Use PCR to
generate an
sgRNA-
encoding
template
In vitro
transcribe
sgRNA
from the
template
Purify and
quantify the
sgRNA via
spin column
Use PCR to
create a
DNA
cleavage
template
Perform
cleavage
reactions
with Cas9-
sgRNA
Analyze
cleavage
efficiency
on an
agarose gel
VI.B
VI.C
VI.D
Guide-it sgRNA In Vitro
Transcription Components v2
Guide-it sgRNA Screening Kit (recommended)
Guide-it IVT
RNA Clean-
Up Kit
Step 1
Step 2
Step 3