Cellartis iPSC rCas9 Electroporation and Single-Cell Cloning System User Manual
(030619)
takarabio.com
Takara Bio USA, Inc.
Page 11 of 24
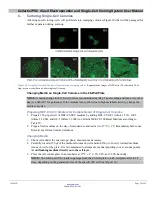
Designing a 56- to 58-nt Forward PCR Primer
The forward (sense) primer must contain the following four sequence elements, in the order
shown in Figure 5, Panel B.
a.
A T7 promoter sequence plus four extra bases (21 total nt) at the 5’ end of the primer.
b.
A transcription initiation site (0–2 guanine (G) residues): The number of Gs added is
dependent on the 5’ end of the target sequence. The T7 promoter requires at least two Gs
for efficient transcription.
NOTE:
If your specific target sequence (see item c, below) already contains two Gs,
there is no need to add extra Gs for transcription initiation. Extra Gs could reduce
cleavage efficiency.
c.
Your specific sgRNA target sequence (20 nt).
d.
The Guide-it Scaffold Template-annealing sequence (15 nt at the 3’end of the primer)
NOTES:
•
A reverse (antisense) primer comes premixed with the Guide-it Scaffold Template.
•
The forward primer should be subjected to salt-free purification following synthesis and
diluted to a concentration of 10 µM in PCR-grade water.
Figure 5. Designing a forward PCR primer to generate a DNA template for an sgRNA containing your target sequence.
Panel
A.
Choose the DNA target sequence that will correspond to your actual sgRNA target sequence.
Panel B.
Design the forward primer
to create the
in vitro
transcription template you will use to generate your sgRNA. [
NOTE:
The T7 promoter sequence (with four extra
bases) and the Scaffold Template-specific sequence do
not
change.]